Machine Learning for Protein-Protein Interactions
 Protein-Protein Interface Featurization
Protein-Protein Interface Featurization
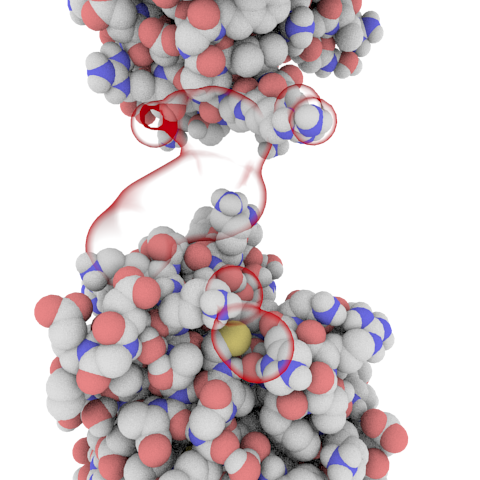
The indentification of biological protein-protein interface is a crucial task of computational biology with applications ranging from drug design to evolutionary studies. The experimental characterization of these interface usually relies on expensive and time consuming methods such X-Ray crystallography. To alleviate this cost a multitude of computational approaches have been developped.
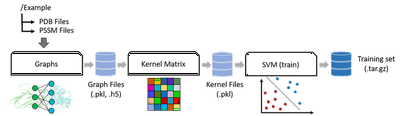
We have recently developped several machine learning based approaches for the classification and scoring of protein-protein interface. Our first attempt, iScore is based on a support vector machine approach using randow walk graph kernels using only about 100 known interfaces in the training set.
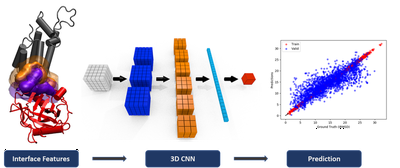
We also explored deep-learning based methods using convolutional neural networks. Featurization of the interface is obtained via gaussian expansion of atomic and residue features on a 3D grid. These models require a large number of training sample that we obtained via molecular dynamics docking simulations on the BM5 dataset.
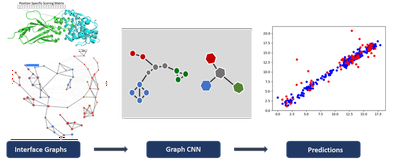
More recently we have explored the possiblity to use graph neural network to encode the strucrture of the interface. This promising approch would remove the need to artificially augment the training data set to account for rotational invariance of the scoring function.